PhD Candidate @ Univeristy of Cambridge & Gates Cambridge Scholar
working on Epigenetics | Bioinformatics | Transposons | Metagenomics | Algae | Chlamydomonas
![]() ORCID iD
ORCID iD


GitHub Portfolio
New repositories
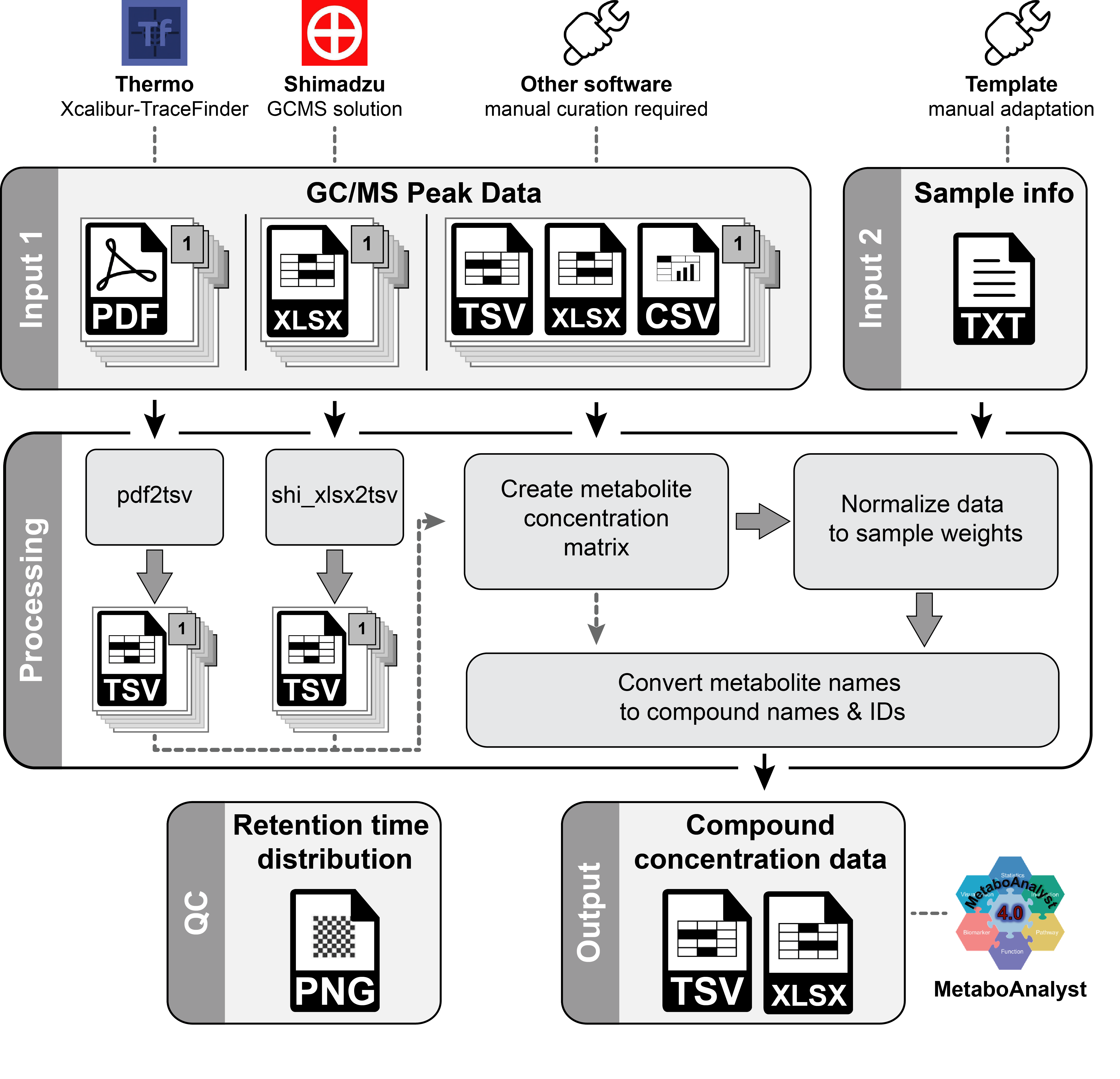
MetaPLMA - Metabolite Peak List Merge & Annotation Tool: A platform independent R tool to merge and annotate metabolite-peak data from GC/MS runs.
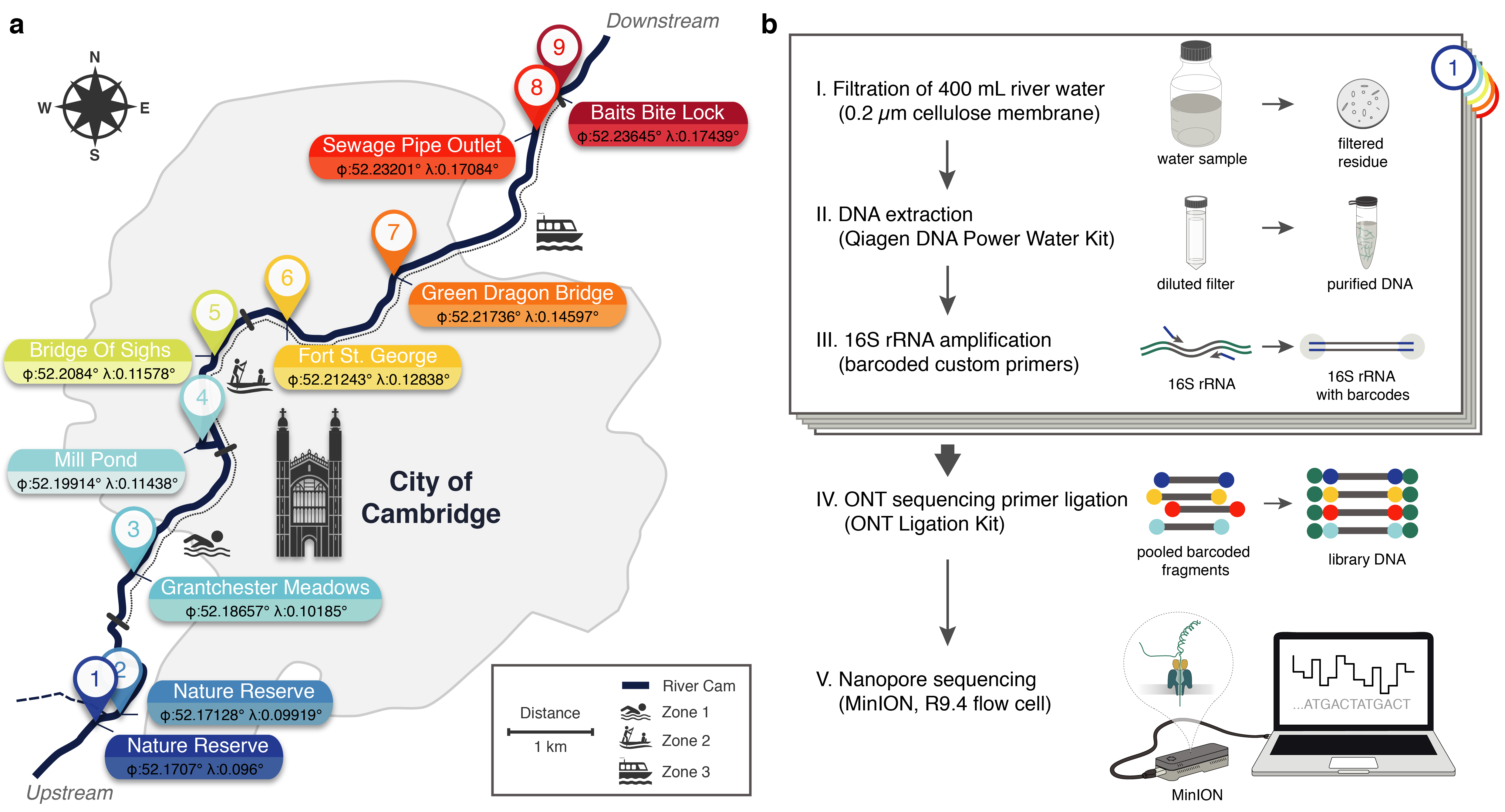
PuntSeq - Chasing the microbial diversity of Cambridge’s freshwater
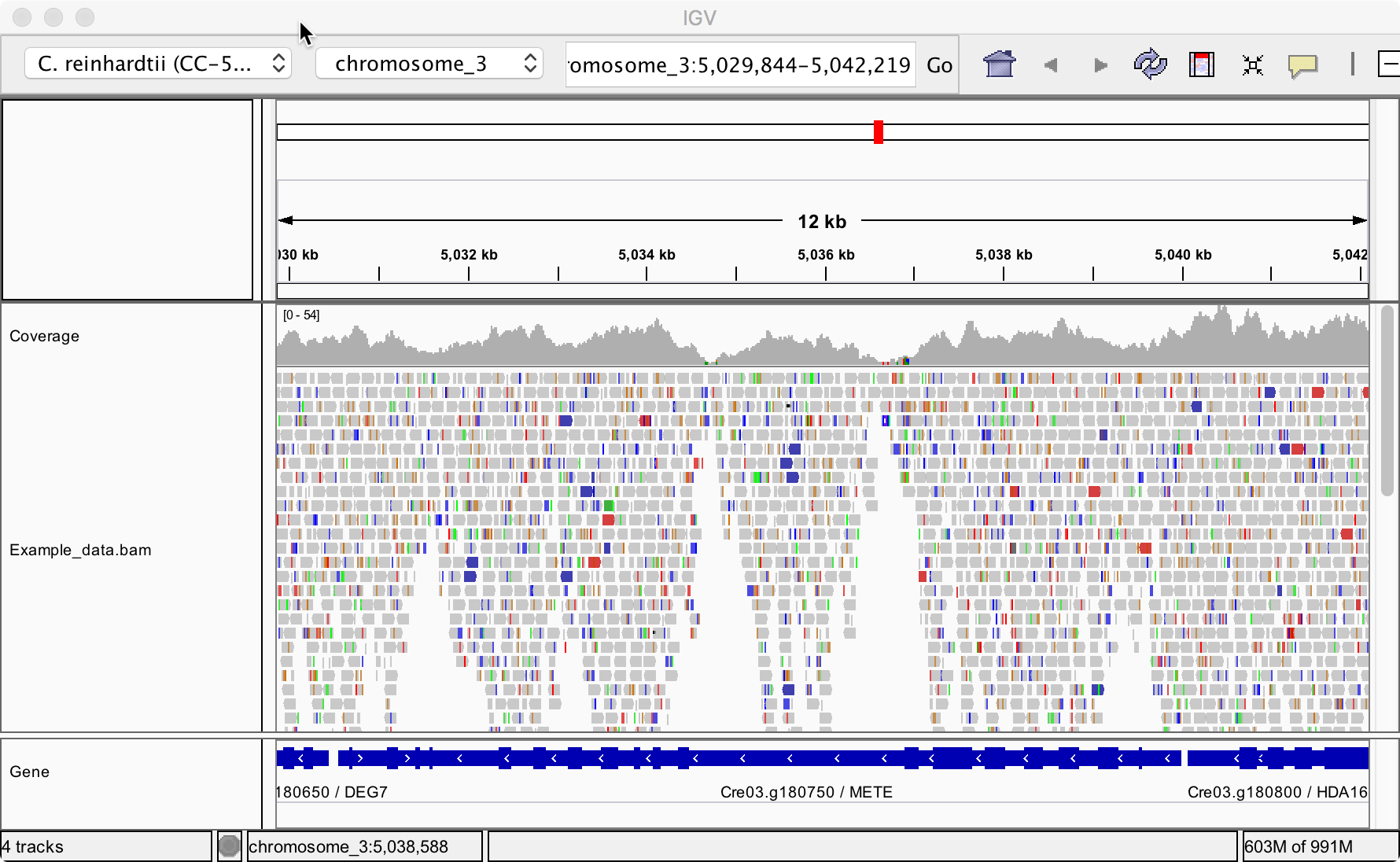
Repository of assembled reference genomes of the green model alga Chlamydomonas reinhardtii including files optimised for visualisation in IGV
Associated Publications
-
Urban L, Holzer A*, Baronas JJ, Hall M, Braeuninger-Weimer P, Scherm MJ, Kunz DJ, Perera SN, Martin-Herranz DE, Tipper ET, Salter SJ, Stammnitz MR (2020) Freshwater monitoring by nanopore sequencing. *BioRxiv preprint 1-38. doi: 10.1101/2020.02.06.936302
-
Bunbury F, Helliwell KH, Mehrshahi P, Davey MP, Salmon D, Holzer A, Smirnoff N, Smith AG (2020) Physiological and molecular responses of a newly evolved auxotroph of Chlamydomonas to B12 deprivation. Plant Physiology 1-20. doi: 10.1101/836635
-
Holzer A (2019). The Chlamydomonas reference genomes for visualisation in IGV (Version v1.0). Zenodo. doi: 10.5281/zenodo.3732212